NewsBlog:
Tentacles test tenets of evolution
Posted by Elie Dolgin
[Entry posted at 18th November 2008 02:42 AM GMT]
Novel genes, rather than regulatory DNA, underlie the evolution of morphological traits, according to research published today (Nov. 17) in PLoS Biology. The new study reports that genes found in simple freshwater animals -- but not in any other evolutionary lineage -- can drive changes in body plan, and stokes the flames of a long-standing debate among evolutionary developmental biologists. "This is the first study that puts together comparative molecular evolution data and experimental data into a cohesive case for this mode of evolution," Günter Wagner, an evolutionary developmental biologist at Yale University, who was not involved in the research, told The Scientist. The underlying genetic basis of morphological adaptation has been hotly debated in the evolutionary developmental (evo-devo) field. On the one side, many argue that innovations in body plans stem from modifications in the spatial and temporal activity of well-conserved regulatory DNA, known as cis elements. Others, however, argue that adaptation and speciation originate from structural mutations in the protein-coding regions of genes themselves. Although many agree that both sorts of changes take place, the relative importance of each process has remained unclear. Now, a team led by Thomas Bosch, an evo-devo biologist at the Christian Albrechts University in Kiel, Germany, has found that expression of a single gene drives major differences in tentacle formation between two closely related freshwater polyps of the genus Hydra -- score one for the structural mutation camp. What's more, the tentacle-related gene was found only in Hydra, with no shared genes in other evolutionary lineages. 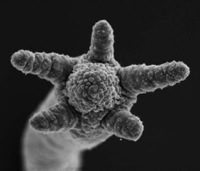 Hydra tentacle formation during budding Bosch's team scanned all the messenger RNAs of two closely related Hydra species for genes differentially expressed in the main polyp-specific structures -- tentacles, nematocysts, and the stalk. Their transcriptome-tracking turned up Hym301, a gene coding for a secreted protein that was expressed in the tentacles of one species and everywhere but the tentacles in the other species. By using transgenic and mutant Hydra that overexpressed Hym301, as well as RNA interference to silence Hym301, they showed that the gene affects the general timing and order in which tentacles arise in the different cnidarians. Genes such as Hym301 that don't resemble known coding sequences in any other organisms -- known as "orphan" genes -- are thought to constitute around 5 to 10% of all genes across most taxonomic ranks, and, yet, geneticists have largely ignored them for two reasons. First, characterizing a gene of no known function is time-consuming and laborious. Second, many suspected that these genes had counterparts in other organisms, but limited data precluded their detection. "You can't ignore [orphan genes] anymore because databases are getting very complete," Bosch told The Scientist. "There are obviously evolutionary selective constraints on keeping them for millions of years... They can't just be nonsense genes that are lying around." Perhaps the best examples of orphan genes, noted Bosch, can be found in the innate immune system where organisms need highly self-specific defense mechanisms. He said he also has unpublished results of Hydra anti-microbial peptide genes with no genetic equivalents outside the genus. Further, he points to human beta-defensin as another lineage-restricted antimicrobial peptide gene that is found only in mammals. "These novel genes are important for adapting an organism to its particular needs," Bosch said. "That's why these genes are not found outside [their] taxon." Caltech evo-devo biologist Eric Davidson, who was not involved in the study, however, doesn't think that the results of the paper can be generalized to account for fundamental evolutionary processes in other organisms. "What makes [a] body plan is fundamentally and generally the deployment of regulatory genes, not the specialized downstream genes," he wrote in an E-mail. Wagner takes a more nuanced stance toward the study's implications, though. "It certainly doesn't undermine the fact that cis-regulatory changes are important in morphological evolution, but it broadens the horizon by showing that other mechanisms, including new genes, can contribute to morphological differences." Image courtesy of PLoS Biology Article originally at The-Scientist.com (free registration, but you have to log in) |





No comments:
Post a Comment